How reliable are the RIBER/DIBER predictions?
RIBER/DIBER predictions are prepared using a classifier trained on all experimental diffraction datasets available in PDB at the time the methods were developed (March 2009 and July 2011 for DIBER and RIBER respectively, data available in the Download section).
DIBER performance
The DIBER server has been recently benchmarked with the structures which have appeared in the PDB since the original version of the program was developed in 2009. Within the error limits, the previously published performance indicators have been confirmed by this analysis. The DIBER classification in combined mode leads to a correct answer for 90% of the protein only, 90% of the DNA only and 80% of protein-DNA crystals.
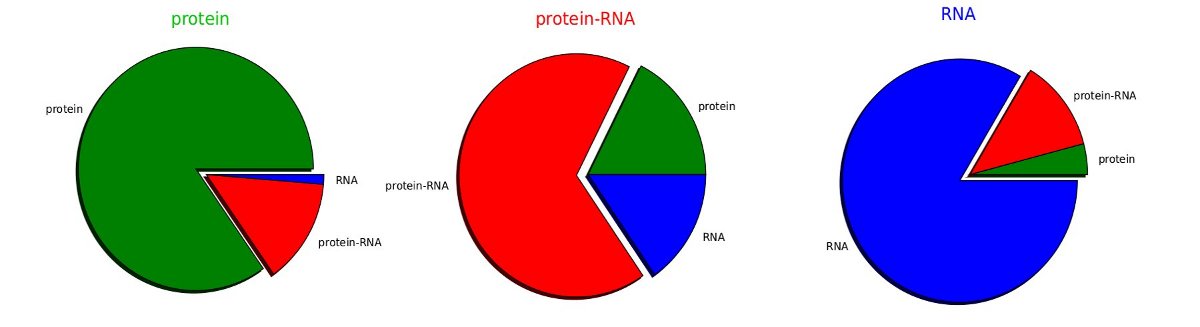
RIBER performance
The RIBER classification performance was estimated using a repeated stratified subsampling validation procedure. Classifier was trained with equal numbers of randomly selected diffraction datasets from each class (RNA only, protein only and protein-RNA complexes). The remaining structures were used for testing. The average classification performance of 100 training and testing cycles was used as an estimate of the true classification performance. The RIBER predictions are estimated to be correct for 80% of the protein only, 80% of the RNA only and 65% of protein-RNA crystals.