Manual (version 5.0 and above)
INTRODUCTION
This is the documentation for RIBER/DIBER version 5 with updated command line interface. The use of this version is strongly recommended. For instructions on how to use older versions refer to the obsolete manual page.
SYNOPSIS
rdiber -mtzin foo.mtz -f F column label -sigf sigF column label -mode diber|riber -phaser phaser output root filename -psout postscript output filename
[Keyworded input]
DESCRIPTION
The program RIBER/DIBER takes diffraction data as input and uses a support
vector machine classifier to predict whether a crystal contains only protein,
only nucleic acid, or a mixture of both. The DIBER is intended to search for
the double stranded B-DNA and not the rarer A-DNA or double stranded RNA. RIBER
was tuned to detect the presence of regular RNA stems in macromolecular crystals.
You can use one of two modes of the program: standalone (DIBER and RIBER)
or combined (DIBER only). In standalone mode prediction is prepared based
on the cube root of the reciprocal unit cell volume of a crystal and the
largest local average of reflection intensities in a thin resolution shell.
The prediction prepared in combined mode is enhanced with the PHASER Maximum
Likelihood Fast Rotation Function score and is more reliable. However, PHASER
is subject to third-party constraints and it is your responsibility that you
comply with any such restrictions. Please contact the PHASER developers to
obtain the necessary permissions for academic and commercial use.
KEYWORDED INPUT
-mtzin mtz filename
Compulsory keyword Input MTZ file containing structure factor amplitudes (F) and its standard deviations (SIGF). Data must extend to at least 3.0 Å resolution.
-mode [diber/riber]
Compulsory keyword Defines the program mode. DIBER should be used if a crystal was grown in the presence of DNA. If a crystalliztion solution contained RNA you should use RIBER.
-f column label
Compulsory keyword Label of column containing structure factor amplitudes (F's).
-sigf column label
Compulsory keyword Label of column containing structure factor standard deviations (SIGF's).
-psout filename
Optional keyword If given, the program will produce graphical output in a postscript format.
-phaser root filename
Optional keyword (DIBER only) If given, the program will run PHASER and use its LLG to enhance prediction reliability. If root filename is not given, DIBER will create its own root name based on MTZ input file name.
OUTPUT DESCRIPTION
The RIBER/DIBER classification of a crystal unknown content can be carried out in two modes: standalone and combined with the PHASER rotation function score. In both cases the text output is basically the same (plain text prediction and related classification probabilities), the graphical output depends on the program mode.
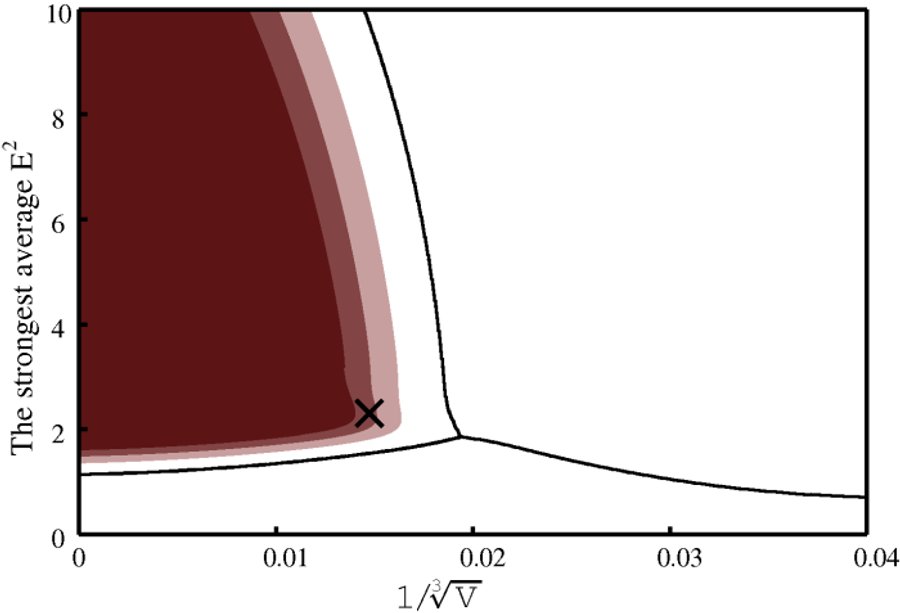
Standalone mode
Two graphs are produced. One presents feature space with the classification boundaries and regions of high level of correct classification probability
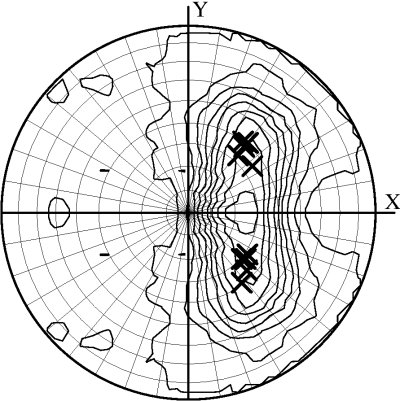
As a by-product of the classification, the program outputs a plot of the local intensity averages on a stereographic net. If a nucleic acid present in your crystal displays long, regular helices their orientations in the unit cell may be derived from the postion of strong signals on the projection.
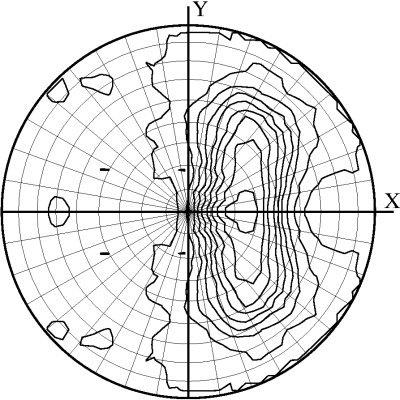
Combined mode (DIBER only)
In this mode, the feature space becomes three-dimensional and thus is not graphically presented. Instead, the top ten solutions of the PHASER rotation function are marked on the stereographic net along with the contour plot of the local intensity averages.